This is a story about the role of data science in accelerating drug discovery with real examples from the leading data-driven biotech and CRO company ZeClinics.
We are Sylvia Dyballa, Technology Director, and Javier Terriente, Chief Innovator Officer and Founder at ZeClinics, and we would like to share how we use artificial intelligence and data science in the biotech industry. But first, allow us to introduce the current challenges in drug discovery.
Table of Contents
- Obstacles in Drug Discovery and Development
- What Is ZeClinics?
- The Zebrafish Model System
- Applications of Data Science in Biotechnology at ZeClinics
- Data Science in Biotechnology: Final Words
- 365 for Business
Obstacles in Drug Discovery and Development
There are over 20 thousand diseases, and most of them have no cure or treatment that can stop the disease progression. Meanwhile, the rate of drug approval by the FDA and EMA is around 50 drugs per year. At this rate, it will take generations to provide treatment for all diseases.
This divergence between the need for more and better drugs and the slow rate of bringing them to the market relates to the lengthy and costly drug development process with a low success rate.
The preclinical phase—i.e., the initial research with in vitro systems, animals, and algorithms to select a candidate molecule for safe testing in humans—takes five years on average, costs approximately $5 million, and has a success rate below 50%. It requires the following converging steps:
- Finding an appropriate therapeutic target—a protein, RNA, or gene related to the disease. Ideally, the pharmacological modulation of this target would allow us to cure or stop the progression of the disease.
- Leveraging this therapeutic target to discover new molecules that can bind and modulate its biological activity. We measure the efficacy and potency of a molecule’s ability to modulate a target—i.e., the concentrations at which it affects the target.
- Ensuring the molecule is safe and not causing side effects (for example, cardiotoxicity) at the efficacy concentration administered. Any side effects should be addressed during the preclinical phase.
- Understanding how well the molecule is absorbed (A), distributed (D) to the tissue or organ of interest, metabolized (M), or degraded, and excreted (E) from the organism. These parameters are called ADME.
The preclinical process is highly iterative, with several screening and chemical optimization rounds. The aim is to select a good clinical candidate—efficacious, safe, and with an adequate ADME profile—for testing in human subjects.
Recent developments in data science and biotechnology help accelerate this process. But before we get to this, let’s introduce ZeClinics.
What Is ZeClinics?
Drug discovery is complex, lengthy, and expensive, and developing new strategies to accelerate the process is mandatory. ZeClinics was born to fulfill this need.
ZeClinics is a biotech company providing preclinical research services to other biotech and pharma organizations. It is also an incubator for spinoff businesses developing new molecules, such as ZeCardio Therapeutics.
The company has vast expertise in developing disease models and understanding the efficacy of new molecules for numerous disease indications. It has also developed innovative assays for understanding drug toxicity in a general or organ-specific manner.
The Zebrafish Model System
The zebrafish has important biological and experimental advantages over other preclinical models. ZeClinics leverages the zebrafish larvae as an innovative experimental model in its research. The main premise is that results obtained from zebrafish translate well to human medicine. Using zebrafish in research has the following advantages:
- The zebrafish provides high biological translatability. It displays an 82% genetic homology to humans for disease-related genes. In addition, its physiology is very similar to that of humans, with a beating heart, a complex brain, and all cell types, tissues, and organs affected by diseases in humans. This allows zebrafish to develop human diseases like Parkinson’s or cardiomyopathies and, therefore, to be useful in drug discovery.
- We can acquire big data. Zebrafish larvae can be assayed at a high experimental throughput, which enables the acquisition of large amounts of data on the impact of drugs or genes in multiple biological processes. We can test dozens of drugs and screen hundreds of zebrafish larvae daily, generating massive amounts of data. This is often a combination of image/video and omics data, processed to reveal the impact of drugs on different physiological functions and processes, such as locomotor and learning behavior, heart and liver function, etc.
Applications of Data Science in Biotechnology at ZeClinics
This combination of biological translatability and big data places zebrafish in a sweet spot for implementing data science (DS) and artificial intelligence (AI) tools. We employ them in various ways, including to automate and accelerate phenotypic and omic analyses and discover new biological paradigms to tackle disease.
Applications of Data Science: Deep Learning for Automated Phenotyping
We continuously generate deep earning (DL) models to conduct case-specific phenotypic data analyses. An emblematic example is ZeCardioAI, a tool that enables the automatic segmentation of the atrium and ventricle of the zebrafish larvae’s hearts in videos.
ZeCardioAI allows us to extrapolate changes in the area from each segmented region and extract—automatically and without human bias—the heartbeat. Then, we translate this into disease-relevant parameters concerning the heart rhythm (BPM, arrhythmias, etc.) and potential contractility defects (chamber size, ejection fraction, strain defects, etc.).
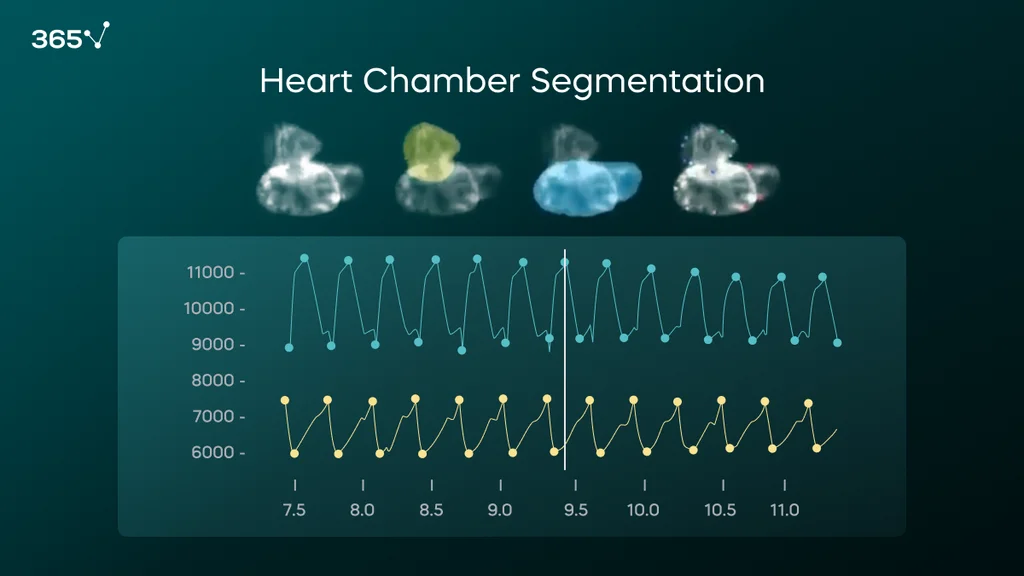
By training a DL model to segment the atrium (yellow) and the ventricle (blue), we can predict those structures in the video and extract heart physiological parameters, such as heart rate, arrhythmias, ejection fraction, etc.
These phenotypes serve to:
- quantify the impact of cardiotoxic drugs,
- analyze cardiomyopathy disease models, and
- validate and discover new therapeutic targets and drugs to treat cardiac diseases.
Another example of the usage of data science in biotech is an AI-powered tool developed by ZeClinics related to developmental toxicity. It helps assess the effect of different molecules on the zebrafish development—i.e., their teratogenic effect. For this purpose, we trained various models on thousands of manually curated images to achieve semantic segmentation of our regions of interest (morphometrics), or image classification.
The following example shows zebrafish larvae image segmentation. Dorsal (top view) and lateral (side view) images are fed to a deep learning architecture. The model was pre-trained on the COCO dataset and fine-tuned on ZeClinics’ data. This project was carried out in collaboration with the Polytechnic University of Barcelona’s (UPC) data science master program.
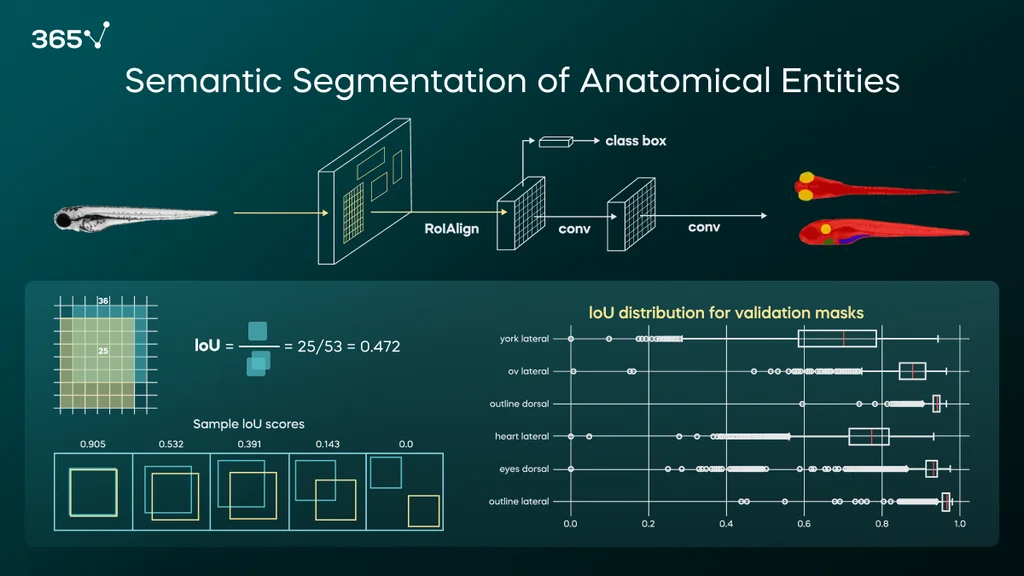
We use the Mask R-CNN architecture to achieve the delineation of anatomical entities in images (e.g., the fish outline in red, the eyes or otic vesicle in yellow, the heart in green, and the yolk in purple). There are multiple ways to assess a model’s accuracy; here, we use Intersection over Union (IoU).
Applications of Data Science: Machine Learning for HTS of Candidate Drugs
Another example of the application of AI in biotechnology at ZeClinics is the use of machine learning (ML) to build classifiers that help predict the compounds’ toxicity when incubating larvae with potentially toxic drugs. This approach assumes that some toxicity indicators are only visible by identifying causalities hidden in large and complex datasets. They are usually inaccessible to the human experimenter without the implementation of advanced mathematical models.
As such, we train our ML algorithms on sets of phenotypes extracted from hundreds of experimental samples with known toxicity. Once trained and validated, these classifiers can predict toxicity for new compounds.
The example below shows how we test whether a drug promotes teratogenicity, i.e., defects in the fetus if exposed to compounds. In this case, we fed dorsal and lateral images of zebrafish larvae incubated with potentially toxic compounds to a ResNet-101–based classifier. The model was pre-trained on the ImageNet dataset and fine-tuned on ZeClinics’ data. Once trained and validated, the model generates a 0–1 confidence score for each phenotype based on which we assign toxicity.
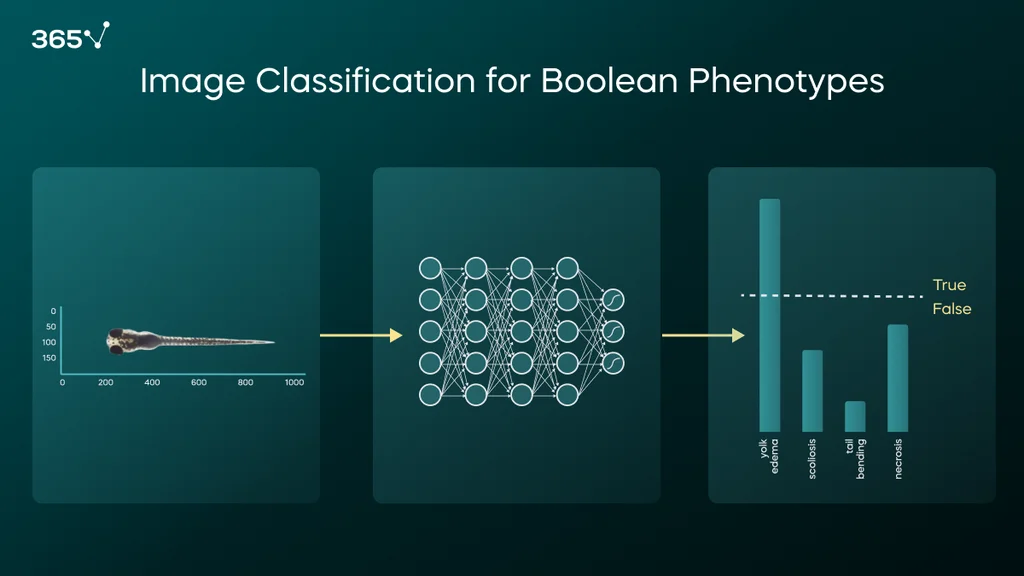
Sometimes, toxicity cannot be determined by quantifying embryonic structures. So, we train a classifier that takes the entire image or a portion of the image as input and outputs a confidence score for the Boolean phenotype for which the model was trained.
Applications of Data Science: Knowledge Graphs for Therapeutic Target Discovery
Another use of artificial intelligence in biotech at ZeClinics is the development of knowledge graphs (KG). KGs are networks composed of nodes (with different labels and properties) and relationships (edges of distinct types and with specific properties). A node in a biomedical KG can be labeled gene/protein, disease/phenotype, compound/drug, etc., while relationships can be labeled induces, activates, cures, etc. So, we can have Protein A → activates Protein B, Gene 1 → is overexpressed in Disease X, and so on.
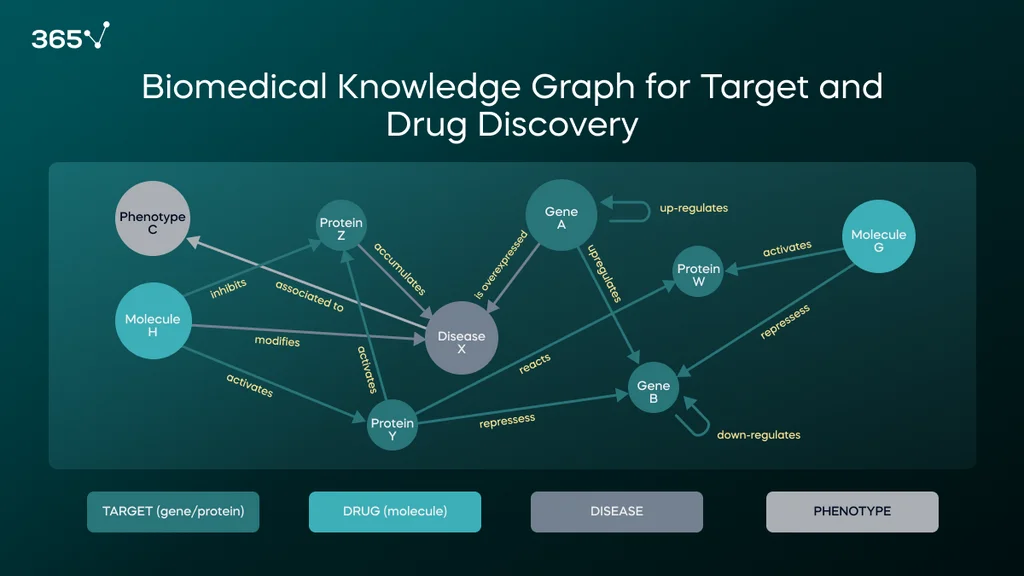
In this example, the nodes include targets (genes or proteins), candidate drugs (molecules), diseases, and phenotypes. The edges are depicted with semantic labels and arrows that show the direction of the relationship.
Knowledge graphs are an elegant way to represent complex systems with many heavily interconnected components. This makes them powerful tools in drug discovery. By using a KG, we can combine public information with our experimental data to gain insights into new targets and potentially important hubs in a disease.
Our DrugDiscovery KG is still under development, but it will comprise a massive network with thousands of nodes and millions of edges containing our understanding of the relationships between targets, drugs, and diseases. This will help us frame the accumulated knowledge from the scientific literature and our research. We aim to identify new therapeutic paradigms, which would be difficult to access via traditional scientific methods, through the combination of external and internal data.
Data Science in Biotechnology: Final Words
ZeClinics was born as a purely experimental research company but soon realized that it had another extremely valuable asset—the wealth of data acquired over the years. We believe that the combination of our experimental data with AI tools and data science can help advance biotechnology and drug discovery.
ZeClinics cannot be described as a purely experimental or digital company. Our activities are multidisciplinary—we operate at the intersection of biology, toxicity, data science, computer science, and, of course, artificial intelligence.
We combine our experimental and digital competencies to generate vast amounts of data in the lab, analyze it, and integrate it efficiently using AI tools. This helps us uncover new biological insights and make better predictions about drug outcomes. Finally, we use our experimental capacities to test our hypotheses’ validity in the lab.
This virtuous cycle combines the best of both worlds and is the best path to discovering new, more efficacious, and safer therapeutics.
365 for Business
Companies from all sectors can benefit from data-driven solutions. But to implement them successfully, they need a data-literate workforce and a data-driven culture.
If you wish to enhance your business performance, optimize operations, and improve outcomes, upskill your employees with data science capabilities. 365 for Business provides numerous data science and analytics courses and live training opportunities for different levels of experience in one learning platform. Request a demo and try it for free.







