Influenza Vaccines and Data Science in Biology
Data science plays a key role in the selection of influenza vaccines. Now, this may sound like an excerpt from a sci-fi novel. But in fact, it is an actual real-life application of modern data science techniques that improves lives today.
In this blog post, we will tell you a fascinating (nerd alert!) story. We are going to talk about viruses (the non-computer kind), about influenza, and how the first vaccine was invented. On top of that, we’ll discuss some data science techniques and tools for analyzing biological data. In addition, we’ll also talk about one of the fundamental visualization techniques for genome data: phylogenetic trees. We’ll see how trees can be implemented when predicting changes in influenza and modeling future behavior of viruses. By the end of this article you’ll even learn about platforms, where you can store and analyze gene data or why not your own genome if you’ve got it.
But let’s take it one step at a time. First, we’ll take a look at what viruses are in general and how they function.
Influenza Vaccines: Flu Mechanism, a.k.a. the Influenza Life Cycle
What are viruses?
Organisms are complicated systems. We are used to the big and visible ones – mammals, birds, reptiles. But there are also microscopic invisible organisms, that actually live among us, or, to be more precise – inside us. Small bacterial or viral cells can infiltrate our bodies down to our cells and make us sick. However, bacteria and viruses are two different types of organisms. And it is a common misconception that the cause of illnessеs is mainly bacteria. Bacteria are a diverse group of cells, but in fact, only 1% of them cause disease. The rest are completely harmless. What about viruses? Well, they are very much the other side of the coin (if that coin was very unfair), as almost all of them (around 99%) cause illness.
And if viruses are so dangerous to other organisms, it certainly makes sense to take a closer look at how they function.
How does a virus function?
The way a virus functions, is by infiltrating a host or host cells. It then uses these cells to replicate and spread through an organism and, well, generally wreak all kinds of havoc. Technically, prior to entering a cell, a virus is called a virion. It’s true that almost everyone uses the term virus to describe both stages. Still, it’s a useful fact you can bring up next time the party conversation is running a bit stale.
Anyway, this is the general mechanism behind how all viruses work. But as you probably guessed, different viruses have specific ways to sneak their way into our cells. And they can vary depending on the virus. Therefore, we’ll see the specifics of how Influenza works in the next paragraphs.
How does influenza work?
You may have heard about an H3N2 or an H1N1 Influenza virus on the news. However, if you’re not a biologist, you’ve probably wondered what these letters and numbers mean. Well, the H stands for hemagglutinin, and N – for neuraminidase. Both H and N are proteins and each of them has its own purpose.
H and N surface proteins -important parts of the influenza life cycle
The H and N proteins sit on the surface of the virus and play a vital role in the influenza life cycle. They aid the penetration of the host cell (Hemagglutinin) and the subsequent replication of the virus (Neuraminidase) in the host cells.
Now, these two proteins can vary a bit in their structure, so different versions of them are identified by a number. An example of that is the H3N2. It contains the third variant of the H protein and second variant of the N protein. As a matter of fact, H3N2 and H1N1 are the two most common subtypes of influenza viruses to infect humans. So, let’s take a look at what their popular names and characteristics.
Hong Kong Flu
H3N2, also known as the Hong Kong flu, caused a pandemic in 1968, which resulted in over a million deaths worldwide. Though not as lethal as the H1N1 strain, it was highly contagious and spread quickly through the population, starting from Asia and later reaching America, via returning troops from Vietnam. By the end of 1969, the virus had reached parts of Africa and South America, as well. 
Spanish Flu
H1N1 was responsible for the swine flu pandemic of 2009, as well as the devastating Spanish flu of 1918. The particular H1N1 strain involved in the Spanish flu was extremely lethal, resulting in over 30 million deaths worldwide. However, the reasons for the high mortality rate remain a mystery. While some scientists suggest an unusually aggressive form of the virus was involved, others claim that the circumstances surrounding the infection: overcrowding of camps and lack of sterile environment during World War I, were the cause for the high death toll.
You’re probably thinking: “If this virus can be so dangerous or potentially lethal, how can we protect ourselves against it? “The answer: influenza vaccines, commonly known as flu shots. So…
What is a vaccine and how does it work?
The first vaccine
The first successful vaccine was introduced in 1796 by Edward Jenner, and it was against the smallpox virus. He observed that people who had previously had a different illness - cowpox, did not contract smallpox. So, if people had the cowpox virus first, they became resistant to the more lethal smallpox. His observations helped create the first successful vaccine. As a result, the smallpox virus has since been eradicated worldwide.
Nowadays, we have different types of vaccines. They aim to help the body’s immune system recognize and prevent a virus from replicating and causing an infection. The process involves using some form of a weakened virus which the immune system can train to recognize. It can then create specific antibodies for it and inactivate it.
Influenza vaccines: what do they contain
Influenza vaccines consist of weakened H1N1 and H3N2 strains. When presented with these, our organism can start creating specific antibodies, which target the virus H1N1 and H3N2 cells. Then when a real virus enters the system, our immune system is prepared and can deactivate it.
Now that we’ve discussed the influenza vaccines and what they contain, let’s take a look at who is responsible for the creation of the vaccine.
Influenza vaccines: manufacturing, selection
WHO decides what the influenza vaccines include. And, no, this was not meant to be a question or a Doctor Who reference, it’s just the acronym for the World Health Organization - WHO and as it happens, they are the people deciding on what the influenza vaccines will contain each year.
But why the need for change?
The reason for an annual flu vaccine: antigenic drift and shift
To answer that, we first need to explain two main mechanisms in viral evolution: antigenic drift and antigenic shift.
Antigenic drift
Imagine you have a group of people, stranded on a raft in the sea. Over time the people on the raft slowly change appearances, they grow a beard, hair gets longer, they get more tanned. In essence, they remain the same people but slightly changed. This is what antigenic drift means, slow changes over time.
Antigenic shift
Now if those people mix their genomes (as none of the kids are calling it) and create a progeny, a.k.a. a child, it will contain a mixture of both their traits. This is what antigenic shift or reassortment means: exchange of genetic material and the creation of a new organism (so a drastic change). In our case, this is a new influenza subtype, such as the H3N1 or H1N1 we talked about earlier.
And that answers our question about vaccine creation and the reason to change it each year. Influenza changes quickly, mutates and transforms. Thus, it’s difficult to find a vaccine to combat all possible circulating influenza virus types.
So, when scientists decide how to formulate the vaccine, they need to choose which strains of the virus to include in order to make it most effective. And the latter depends on how closely the vaccine resembles the influenza viruses, which will dominate during the upcoming flu season.
Predicting the influenza spread – data science
How to predict upcoming influenza virus types?
This is where data science comes into play. Based on existing data about former and current virus spread and variants, scientists try to model and predict the future behavior of viruses, using machine learning algorithms.
In order to do that they first need an appropriate way to handle information about viruses, or more precisely their genomes. This is done via analysis of genetic data. But what is genetic data, exactly?
What are Genomes and Gene Data?
Genetic data includes the genome of organisms or parts of it. It’s usually comprised of DNA, represented in the form of strings. In the case of Influenza, it contains RNA, which is what some viruses have as their genetic material.
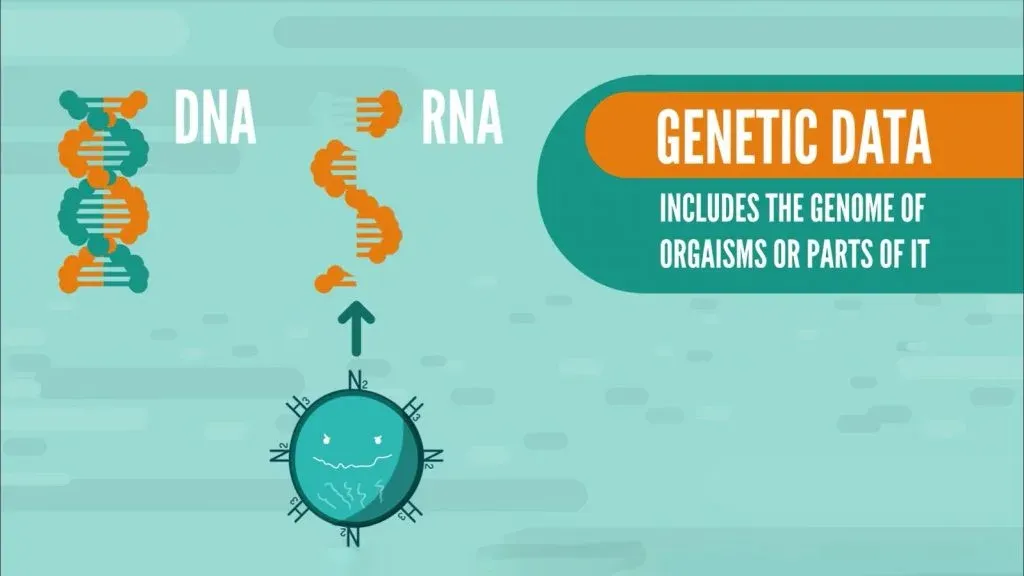
Once we have our data, it’s time to think about how we can make sense of it, which means we first need a way to visualize it.
There are quite a few options. However, we’ll focus on one in particular, which is a staple: the phylogenetic tree.
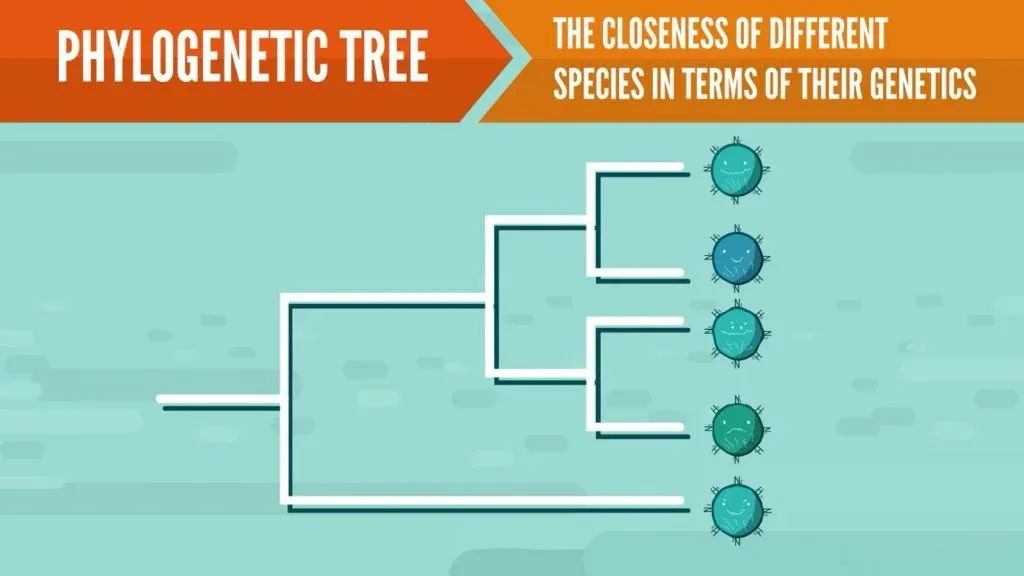
Visualization techniques: Phylogenetic trees
Phylogenetic trees, also known as evolutionary trees, represent the closeness of different species in terms of their genetics. Basically, they are a diagram showing evolutionary relationships between species. In the case of influenza, such trees can be used to visualize different strains of the virus.
Prediction models
It’s time to put all of this together and get to the final point. Namely, prediction using machine learning techniques.
Imagine you already have your biological data in the form of Influenza genomes or antibodies and have represented it using trees. Using the information obtained from the trees, you can employ different machine learning techniques to model future behavior or spread of the Influenza virus.
These include the use of non-negative least squares, construction of maximum likelihood trees or the use of scoring methods. Examples of the latter include the construction of similarity classes and substitution matrices to explain antigenic differences of viruses. And we’ll provide an overview of a few different techniques in the next paragraphs.
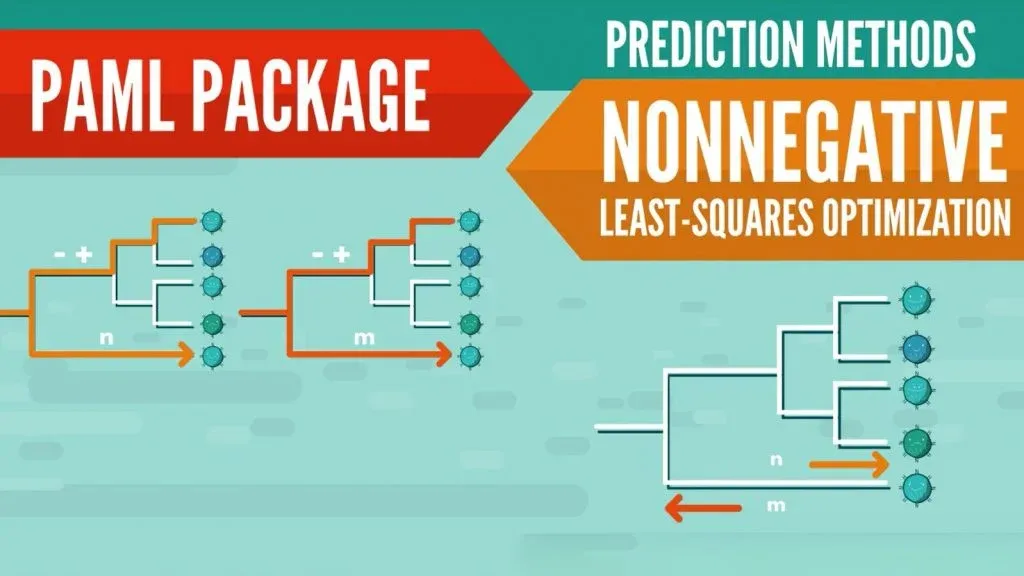
Nonnegative least squares
One common method for prediction is introduced by Steinbrück and McHardy. It uses a nonnegative least-squares optimization, which measures distances between branches of a phylogenetic tree. They use a bidirectional weighted phylogenetic tree and determine sets of coding changes on the surface of the H protein. The model can then identify the antigenic impact of different influenza strains.
Phylogenetic Analysis by Maximum Likelihood or PAML
Another way to perform phylogenetic analyses is to use the PAML package, which contains programs for phylogenetic analyses of genetic data using maximum likelihood (ML). The way this is done is by taking a set of trees and evaluating their log-likelihood values under different models. These models estimate some parameters while allowing for others to vary. This way they can incorporate the variety of gene types in influenza strains and their surface H protein.
Comparison of a Tree-based and Substitution Model
The third method worth mentioning is… Well, actually there are two different methods: one tree-based and a substitution model, as well as a comparison between the two. It feels like we tricked you, but we promise it makes sense to talk about those two in particular.
This last approach to predicting influenza strains was proposed by Neher et al. It includes a tree-based model, which has a test and a reference influenza strain and creates a weighted phylogenetic tree. The substitution model uses sums of contributions associated with amino acid substitutions between reference and test viruses. Through data collected between 2002 and 2015, Neher et al. demonstrated that both the tree-based model and the substitution model perform similarly in terms of prediction accuracy.
Choosing the “best” method (in data science)
We did say the last two models were mentioned with a specific purpose. And it was to illustrate a very common problem in data science: Machine Learning provides a large variety of tools which allows us to analyze data and make prediction models. In some cases, especially if you are a novice in the field, this can become overwhelming. In the case of Neher, we see two distinct techniques yielding similar result on the same problem. This can often be the case in practice: two or more algorithms perform similarly well on a given data set. The choice of the “right” algorithm then can depend on the specifics of the task we’re given or be determined by other factors (speed, scalability, interpretability of the model, the list goes on).
This is also known as the “No free lunch theorem”, a common problem in machine learning, stating there is no one model to solve all problems. An important part of the job of data scientists is to know the strength and weaknesses of each method and always choose an appropriate tool to solve the problem at hand.
Genomes, the up and coming field in data science
And that pretty much brings this article to a close.
That was quite of a roller coaster, right? We started from learning about the flu and how a virus works and went through the history of the first vaccine and the biggest flu pandemics. And the time when we talked about antigenic shifts and drifts? We had a lot of fun explaining these in particular.
We also discussed different types of biological data and their visualization. Finally, we learned how to make predictions using different machine learning techniques.
In conclusion, data science is not just a tool used in the IT Domain or by large corporations. In fact, it plays an increasingly important role in (life) sciences. Moreover, medical and biological applications are becoming increasingly important and widespread. In fact, big tech companies like Google and Amazon started their own genome projects recently, allowing users to store and analyze their own genome on their respective cloud platforms. Microsoft entered the field as well with the release of Microsoft genomics on their Azure cloud.
And if they’re doing it, it’s a safe bet to assume that genomes and their analytics using machine learning are worth looking into. The way things are running, genomes and their analytics might soon become part of our everyday lives. So, we believe it makes perfect sense to get acquainted with the field. And, after the introduction we just gave you, we’re sure you’ll have no trouble doing that.
Start your data science journey with our Introduction to Data and Data Science course. Learn all you need to launch a successful career in the field with our beginner-friendly courses and practical exercises. Sign up for free and try the 365 Data Science learning platform now.







